FPWR collaborates with University of Connecticut-Wesleyan University Stem Cell Core to create a centralized high-quality PWS biobank of induced-pluripotent stem cells (iPSCs).
The Foundation for Prader-Willi Research (FPWR) has established a partnership with the University of Connecticut-Wesleyan University Stem Cell Core to develop a centralized high-quality biobank of iPSC lines derived from individuals with Prader-Willi syndrome (PWS). The PWS iPSC Biobank maintains a high quality-controlled iPSC lines generated from donors with PWS. These lines are available for academia and industry worldwide.
In collaboration with Dr. S. Chamberlain and the UConn Stem Cell Core in Farmington, CT, we have established a validation and quality control (QC) package that has been made available to characterize iPSCs that have been identified as having high profile and value to our research community. This collaboration seeks to provide material containing the most pertinent available information using consistent and high-quality characterization methods and facilitate establishment of the iPSCs in the recipient laboratory. Each PWS iPSC sample provided through the bank has undergone a select set of validation assays. Assays have been performed in the presence of a complete set of comparator or control iPSCs maintained in Dr. Chamberlain’s Laboratory and the Stem Cell Core. The characterization package includes:
- Microbiology Screening - cultures are screened for presence of microbiological contamination, including mycoplasma, bacteria, fungi, and human bloodborne pathogens.
- Culture quality characteristic assessment report - includes morphology, proliferation, and attachment, viability from cryopreservation and recovery, and proclivity towards random differentiation during maintenance culture.
- Cyto-SNP Analysis (Illumina) - This is a whole-genome scanning panel designed for efficient, high-throughput analysis of genetic and structural variations that are relevant to human diseases, including duplications, deletions, amplifications, copy-neutral loss of heterozygosity (LOH), and mosaicism.
- Pluripotency Test and Lineage Score (Taqman Scorecard using undifferentiated and randomly differentiated iPSCs) - The Scorecard Panel is a comprehensive gene expression real-time PCR array comprised of a combination of control, housekeeping, self-renewal, and lineage-specific gene detection reactions. The resulting expression analysis report is generated by comparing the expression pattern of the test iPSCs (undifferentiated and randomly differentiated) against a reference standard composed of multiple functionally validated cell lines. The data is used to confirm signatures of self-renewal, terminally differentiates states, (i.e. in samples that have been randomly differentiated in vitro, and if the starting material is pluripotent, will be expected to develop into tissues of multiple cell types) and the data can be used to infer sample differentiation potential. Reference: https://doi.org/10.1007/7651_2014_109
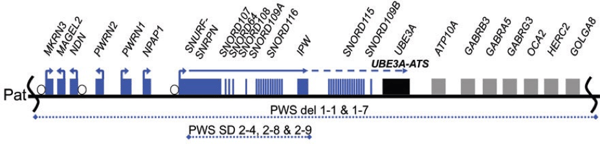
- PWS-associated Gene Expression for MKRN3, MAGEL2, NDN, SNRPN, SNORD116, and IPW genes - qRT-PCR is used to assess expression of these genes in the iPSCs. These imprinted genes located in the 15q11-q13 region provide evidence of how these iPSCs reflect gene expression patterns expected from individuals/tissues/animal models with PWS.
- DNA Methylation Assay (PWS-Imprinting Center; PWS-IC) - PWS is an imprinting disorder characterized by the differential methylation that marks the PWS-IC. This assay is used to confirm that the appropriate methylation imprint is maintained in the iPSCs.
The results of these tests are publicly available and linked below. Publications that may result from the use of the PWS iPSC biobank should cite the validation process performed by the UConn Stem Cell Core in collaboration with FPWR.
Cell lines currently available at PWS iPSC Biobank
While the PWS iPSC biobank is expanding, three human iPSC lines are currently available for distribution including:
PWS large deletion line 1.7 (PWS1.7)
PWS patient fibroblasts with a deletion of paternal 15q11–q13 (see diagram below) were reprogrammed into iPSCs using a mixture of retroviral vectors encoding OCT4, SOX2, KLF4, MYC, and LIN28 and intially characterized in Chamberlain et al., PNAS 2010. This line was further characterized alongside the other lines below in each of the linked publications and validation/QC reports.
PWS small atypical deletion line 2.9 (PWS2.9)
PWS patient fibroblasts with a 187 kb microdeletion within paternal 15q11–q13 (see diagram below) were reprogrammed into iPSCs using a polycistronic STEMCCA lentiviral vector encoding OCT4, KLF4, SOX2 and CMYC and initially characterized in Martins-Taylor et al., Hum. Mol. Genet. 2014. Results of the additional validation and QC described above can be downloaded here.
PWS maternal uniparental disomy line 1.2 (PWSUPD1.2)
The initial generation and characterization of this iPSC line from a PWS UPD patient is described in Langouet et al., Hum. Mol. Genet. 2018. Results of the additional validation and QC described above can be downloaded here.
How to obtain cells from the FPWR PWS iPSC Biobank
Investigators interested in obtaining these cells should visit https://health.uconn.edu/stem-cell-core/services/distribution-of-human-pluripotent-stem-cell-lines/ or contact the UConn Stem Cell core at ucscicore@uchc.edu.
For any additional information please contact Theresa Strong, PhD at theresa.strong@fpwr.org.




